Mixed Infection
Mixed infection, where more than one distinct strain of a pathogen is present in a host at the same time, can be a relatively common occurrence in bacterial and viral infection. In TB for example, up to 20% of clinical samples have been estimated to be mixed.
Identifying these complex infections can be important clinically as these samples may be hetero-resistant and the minor frequency strains may be transmitting. Importantly, failing to account for potential mixed infections can lead to a single erroneous consensus sequence that, causing issues with downstream phylogenetic and genomic analysis.
This practical will walk you through the main steps to identify mixed infection from allele frequencies from VCF files produced from short-read sequence data. We will you my tool, MixInfect2, which is freely available in R from the GitHub, or using ‘devtools’ as we will see in the practical. Please read the paper if you would like more information about how the tool works.
We will the following data for this exercise:
SNPs_filtered.vcf - The multi-sample VCF file produced in practical session 2 that contains SNP variant calls from 3 M. tuberculosis samples.
MaskedRegions.csv - A .CSV file with genomic features that we want to mask from the mixed infection analysis as variation in these genes can obscure the signal from mixed infection.
1. First we will set up the MixInfect2 function from the MixInfect2.R script in your folder:
# Set packages
source("MixInfect2.R")2. Next, we can run MixInfect2. This will take the VCF file and the CSV file with masked regions as input
results <- MixInfect2("SNPs_filtered.vcf",maskFile = "MaskedRegions.csv",prefix = "TB_SNPs")3. This will save the results of MixInfect2 to the results variable. View the results:
head(results)4. We can see that we have some potential mixed infection in our three samples. Below are two allele frequency plots from our samples.
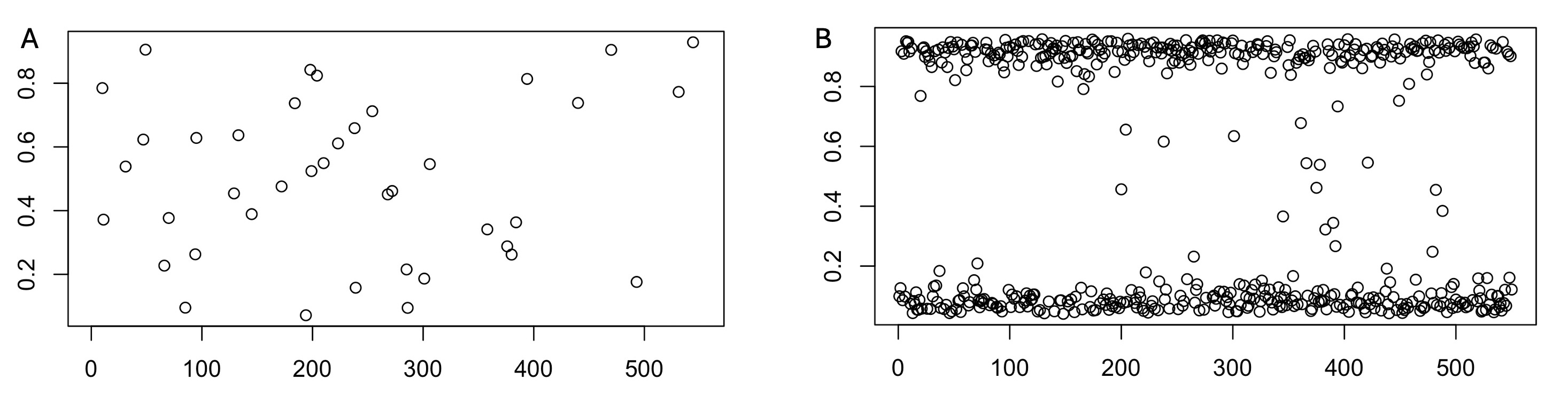
Discussion: Which of our samples do you think these plots may belong to?
You can use these results to remove putative mixed infection from your analysis, or you can potentially reconstruct the constituent sequences of mixed infection using the reconstructConstituents.R script in MixInfect2, though there are limitations in this. Please see the associated paper for more detail on reconstructing mixed constituent sequences.