Inspecting the temporality of sequences using TempEst
Before building a timed phylogeny, it can be important to test for temporality in your data. Temporality in this context refers to the presence of a molecular clock signal, meaning that genetic changes accumulate at a roughly consistent rate over time. This is essential for reliably estimating divergence times in a phylogenetic tree. This analysis can inform the clock model to select when building a timed phylogeny or identify sequences that may be problematic, such as those with incorrect dates or significant rate variation.
Tools like TempEst can be used to plot the root-to-tip distances for each sequence from an un-timed phylogeny against sequence dates. In a dataset where a molecular clock holds, you would expect a positive correlation between these distances and time. The root-to-tip distance in a phylogenetic tree is the evolutionary distance from the root of the tree (representing the most recent common ancestor of all the sequences in the tree) to the tips (representing the observed sequences).
The data we will be using in this exercise are:
-
TB_cluster_ML.tree – A Maximum Likelihood phylogeny from 37 M. tuberculosis samples collected between 2005 - 2014 in British Columbia. This is an untimed phylogeny so the branches will be in units of substitutions/site. This tree has been rooted by an outgroup sequence that has been removed.
-
TB_cluster.txt – A text file with two columns, the name of the 37 M. tuberculosis samples and their collection dates.
Open TempEst:

1. You will be prompted to open a file, select the TB_cluster_ML.tree file. This will open the following screen:
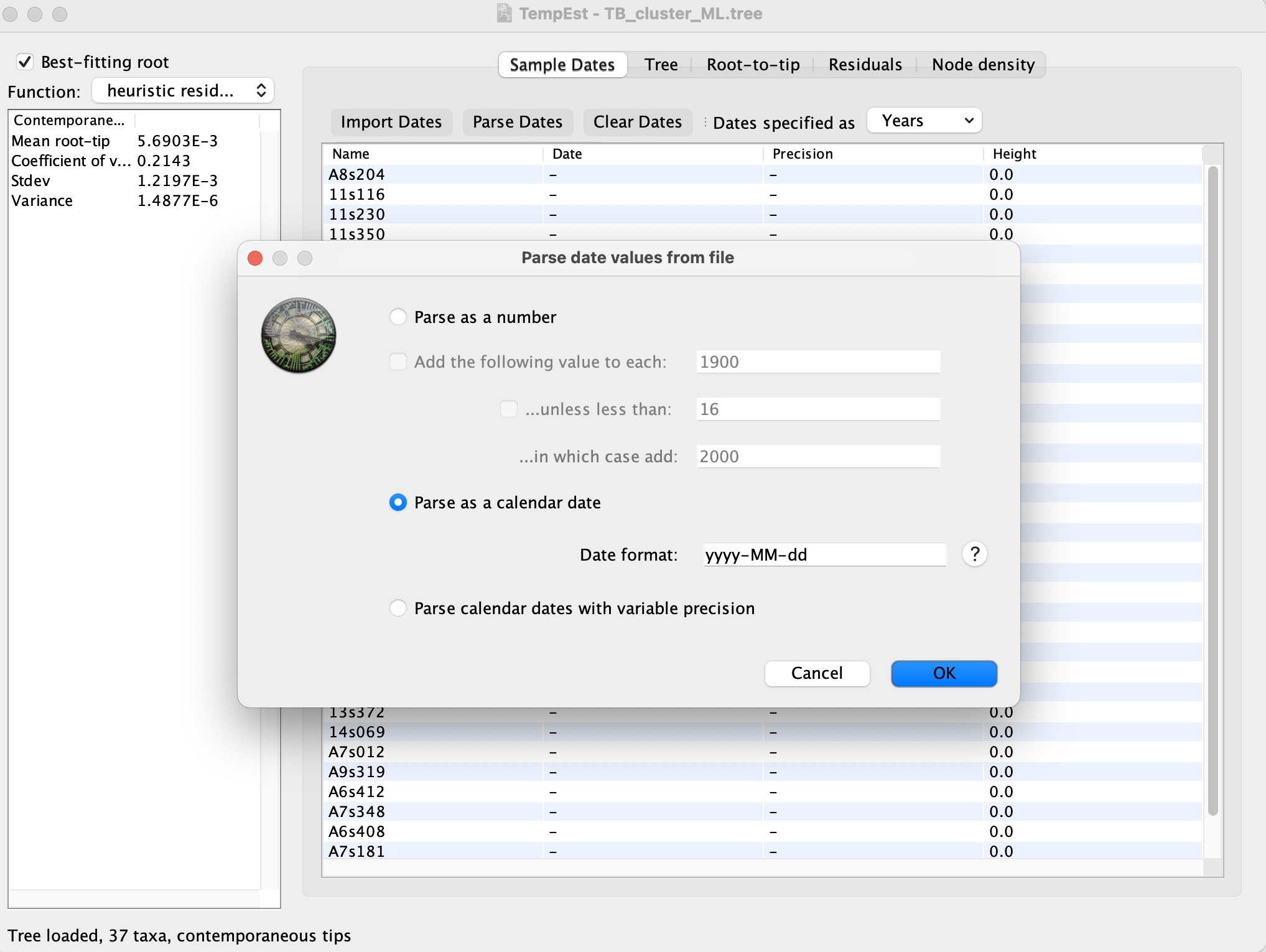
2. Select Import Dates and open the “TB_cluster.txt” file to import the dates. This will prompt the following screen to parse the dates in the correct format. The dates are in the “yyyy-MM-dd” format:
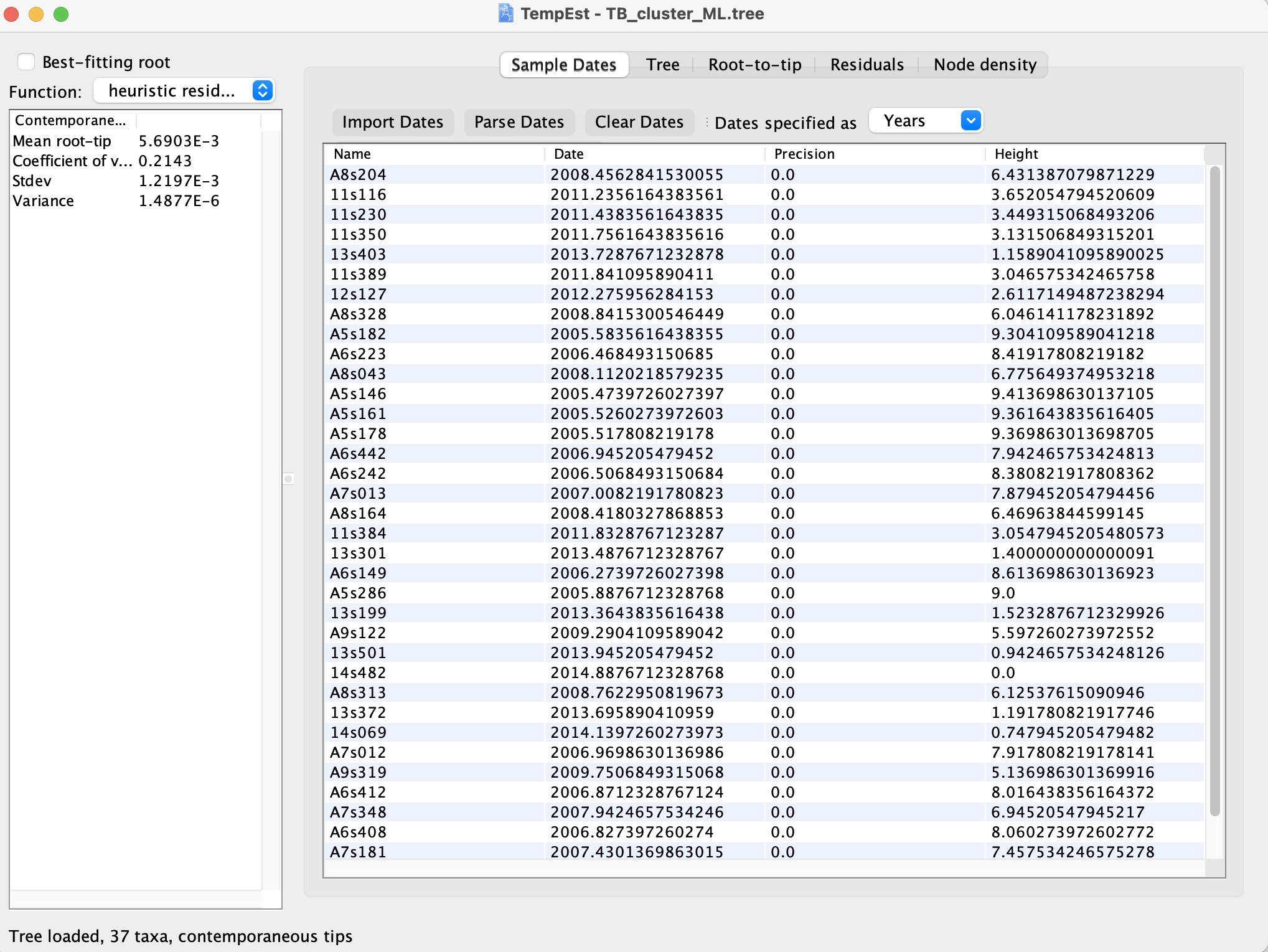
3. You should see the following screen showing that the dates have been loaded correctly:
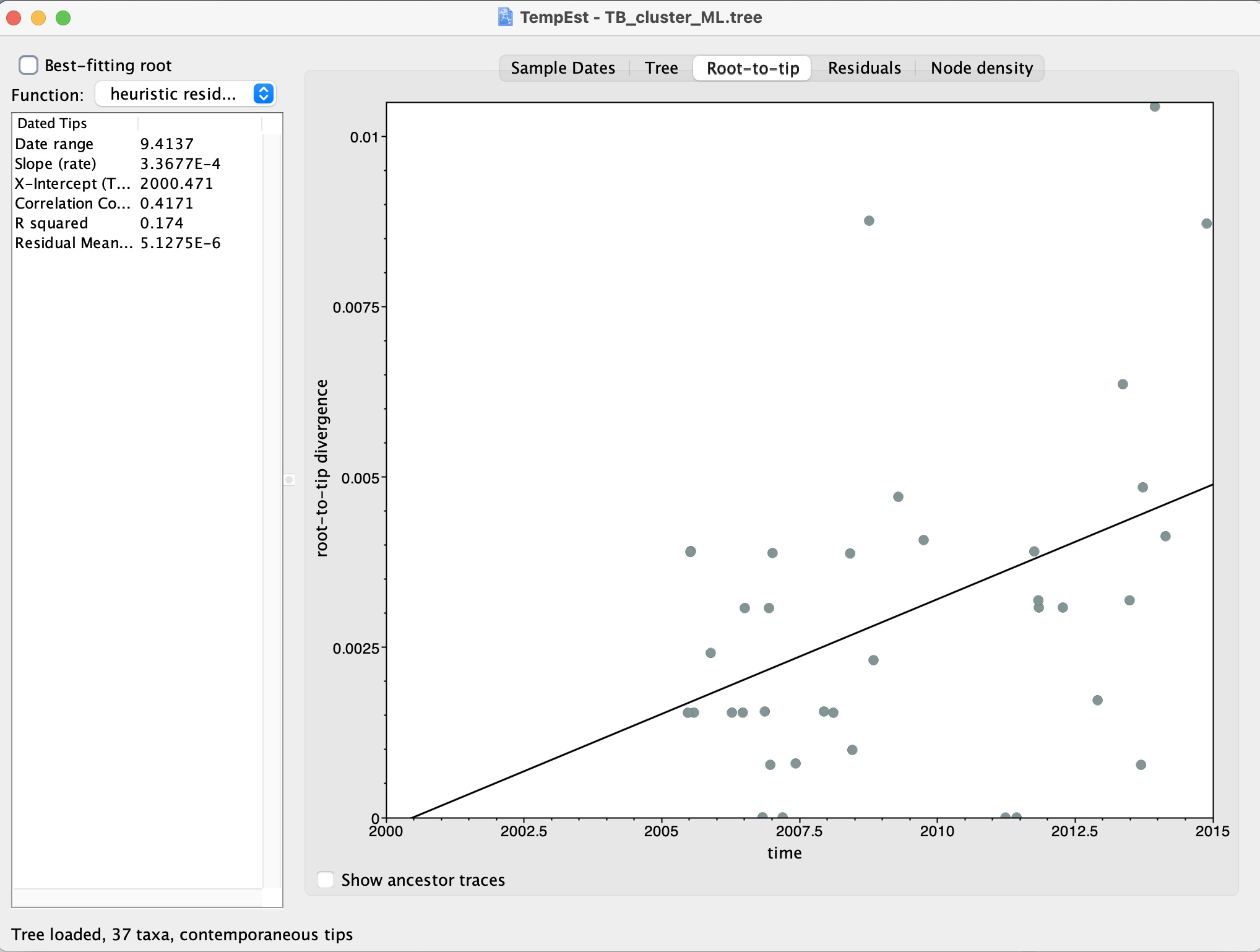
4. Finally, we can click on the ‘Root-to-tip’ tab. This brings up a plot of the root-to-tip distances against the sequence dates. We should see a positive correlation if there is temporality in the data:
If your analysis shows a good temporal signal, you can proceed with more confidence to build a timed phylogeny using software like BEAST, which can estimate divergence times and evolutionary rates.
If there’s no clear temporal signal, consider reviewing your data and methods. It might be necessary to exclude problematic sequences, refine your sampling strategy, or reevaluate the assumption of a molecular clock for your dataset.
Questions to discuss:
What is the correlation in our data?
Do you think there is temporality?
What can be do to increase the temporal signal in our dataset?
If we cannot increase the temporality, can we still build a timed phylogeny?